Discrete Time Models Using ctsem?
The ctsem marketing department seems to have done a poor job getting out the message that all of the dynamic systems stuff of ctsem – fluctuating / coupled latent processes over time, intervention effects, higher order models, simple and complicated forms of heterogeneity across subjects and or time, latent interactions / state dependent parameters – are also possible in the computationally simpler discrete time (e.g. vector autoregression / structural equation model) setting. Whether this is adviseable, or maybe just a neat stepping stone, depends on how you think about your constructs of interest and what you want to infer about them. For some elaboration on the treatment of time, consider https://osf.io/xdf72/
For this example, we have collected data from workers starting a new job. The data include measures of stress, and quality of the employees output. We are interested in a) how stress and quality change from the beginning of the job, b) whether there is any evidence for one process causing the other, and c) to what extent there are individual differences in the start and long term levels of each, and if / how the individual differences are related. Disclaimer: This is a very limited / stylised / toy example to show some of the ctsem basics, for real problems please do much more.
The following block loads the ctsem library then generates the data – interesting for some but less interesting for others so perhaps skip!
set.seed(1)
library(ctsem)
#generating data - can ignore
set.seed(3)
gm <- ctModel(LAMBDA=matrix(c(
1,0,
.8,0,
0,1),ncol=2),
MANIFESTVAR = diag(.5,3),
DIFFUSION=matrix(c(.8,.5,0,.2),2,2),
DRIFT=c(
-.5,0,
-.3,-.2),
T0MEANS = c(20,-10),
TRAITVAR=matrix(c(1,.7,0,.2),2,2),
Tpoints=10,manifestNames = c('Stress','Stress2','Quality'))
## [,1]
## [1,] "20"
## [2,] "-10"
## [,1] [,2]
## [1,] "-0.5" "0"
## [2,] "-0.3" "-0.2"
dat <-data.frame(ctGenerate(ctmodelobj = gm,n.subjects = 100,burnin = 2))
head(dat)
## id time Stress Stress2 Quality
## 3 1 0 6.6351900 0.3682511 -8.475775
## 4 1 1 2.6242786 0.6501790 -11.158248
## 5 1 2 -0.3425943 0.3968806 -11.637336
## 6 1 3 -1.2786638 0.8494424 -12.036826
## 7 1 4 -1.1674515 -1.1327005 -10.259180
## 8 1 5 -1.6114791 -0.2277730 -10.375412Model Specification
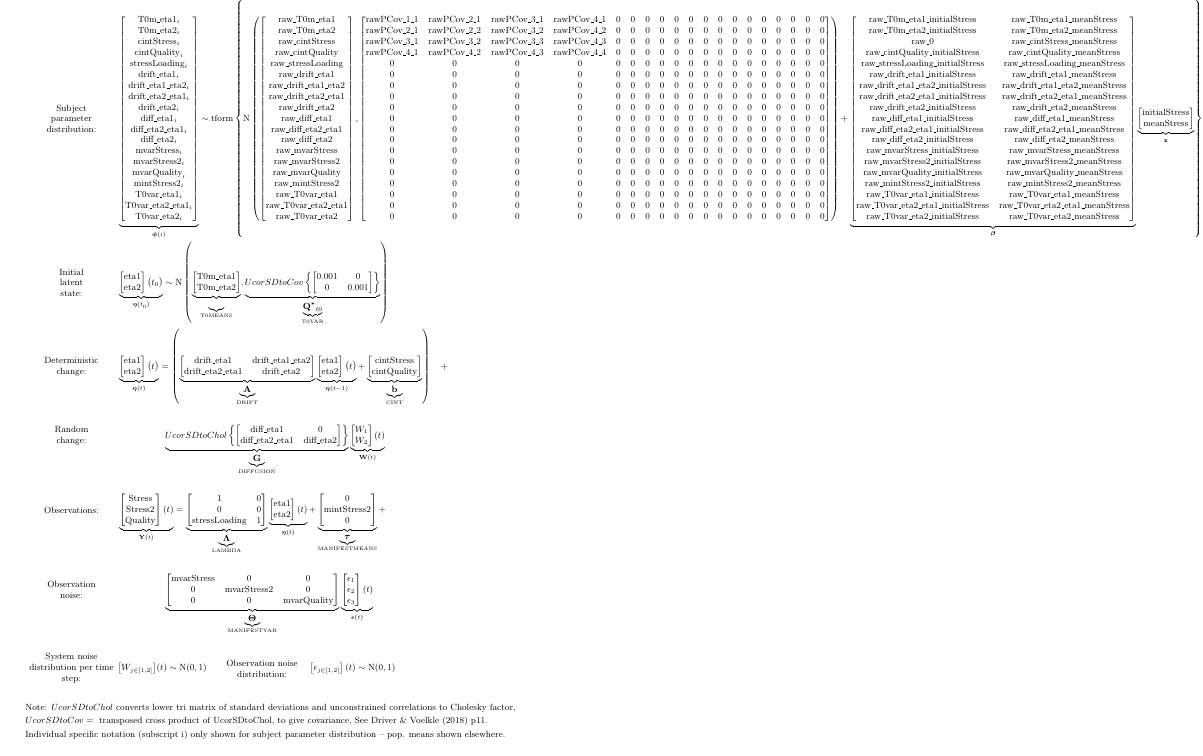
We specify a bivariate process model, with each process measured by a single (noisy) indicator (leaving out our second stress indicator for the time being). Intercepts (here, MANIFESTMEANS) and initial parameters (T0MEANS) default to indivdually varying, but we specify explicitly here in any case as a demonstration (using the | separator notation). In fact, we specify the entire model explicitly, while since most parameters here are freely estimated, we could have left it to ctsem defaults.
model <- ctModel(
LAMBDA = diag(2), #factor loadings / measurement structure
type = 'standt', #could also choose stanct for continuous time modelling.
manifestNames = c('Stress','Quality'),
latentNames = c('Stress','Quality'),
T0MEANS = c('t0_stress','t0_quality'), #starting values for latent processes
MANIFESTMEANS = c('mi_stress||TRUE','mi_quality||TRUE'), #intercepts on observed variables
MANIFESTVAR = c( #vectors are coerced rowwise into matrices of the right shape, 2x2 in this case
'merr_stress',0,
0,'merr_quality'),
DRIFT = c( #temporal dependence matrix, autoregression on the diagonal, cross regression off diagonal
'a11','a12',
'a21','a22'),
DIFFUSION = c( #system noise. sd's on diagonal, unconstrained correlations on lower triangle
'diff11',0,
'diff21','diff22'),
CINT = 0 #single values are repeated as many times as necessary (here, twice, one for each process)
)
#ctModelLatex(model) #view model in pdf (i.e. readable) form. Requires latex compiler
Model Fit and Plots
fit <- ctStanFit(datalong = dat,ctstanmodel = model, cores=2)
#ctModelLatex(fit) #view dynamic system estimates in equation form, requires latex compiler
ctKalman(fit,kalmanvec=c('y','yprior'),plot=TRUE, subjects=1:2) #forward predictions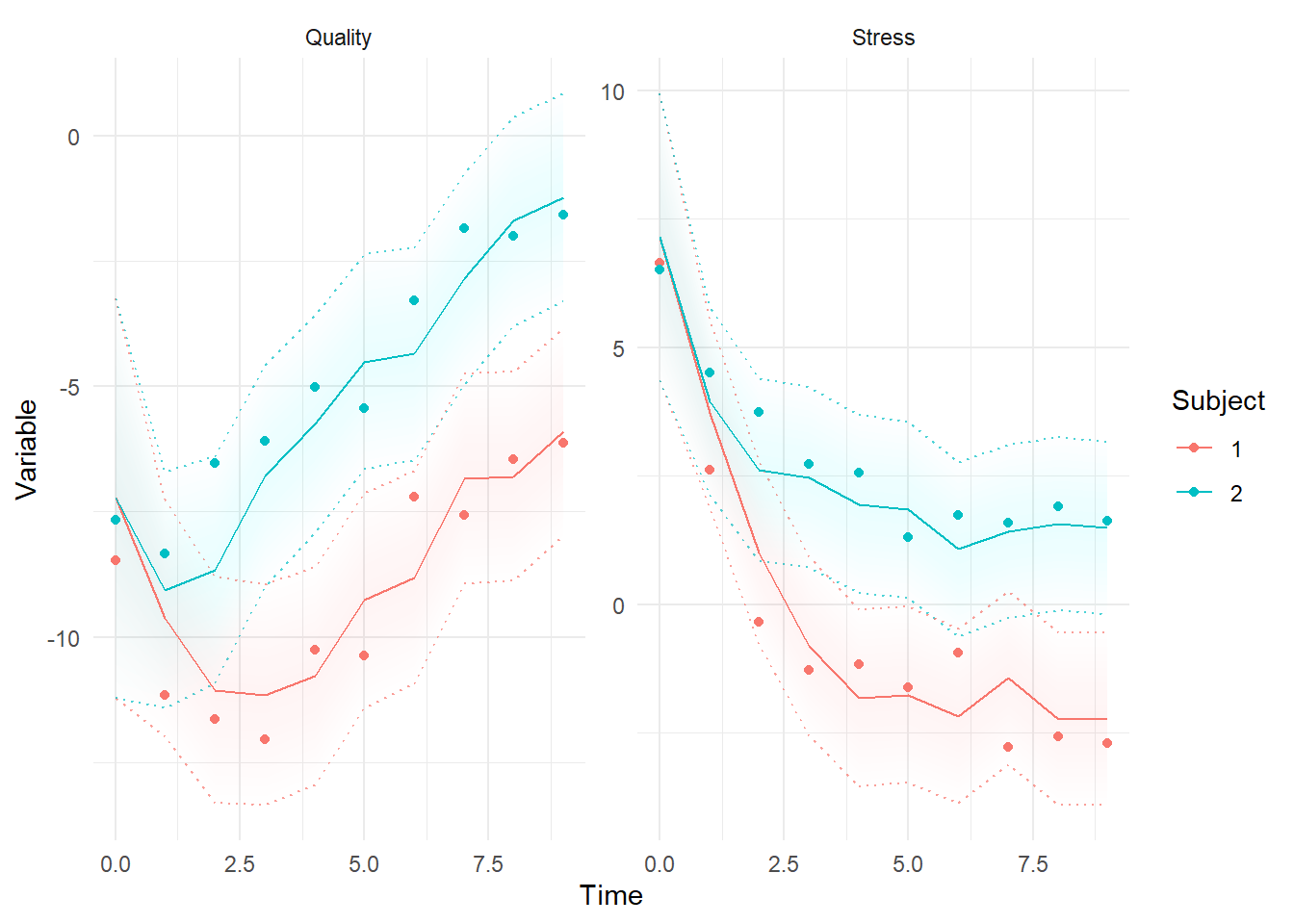
ctKalman(fit,kalmanvec=c('y','ysmooth'),plot=TRUE, subjects=1:2) #best estimates conditional on all data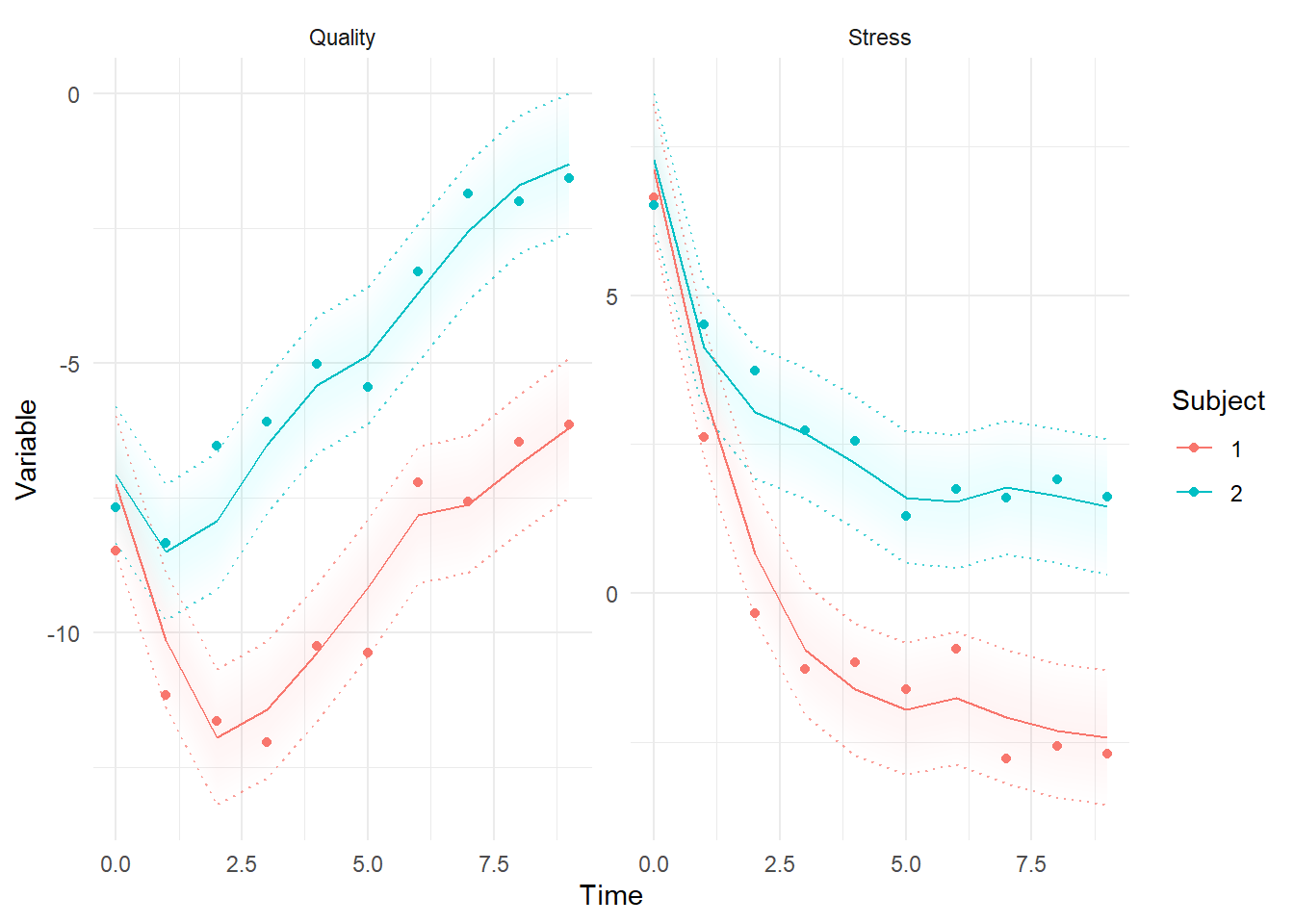
ctKalman(fit,kalmanvec=c('y','yprior'),plot=TRUE, subjects=1,removeObs = T) #predictions before seeing any data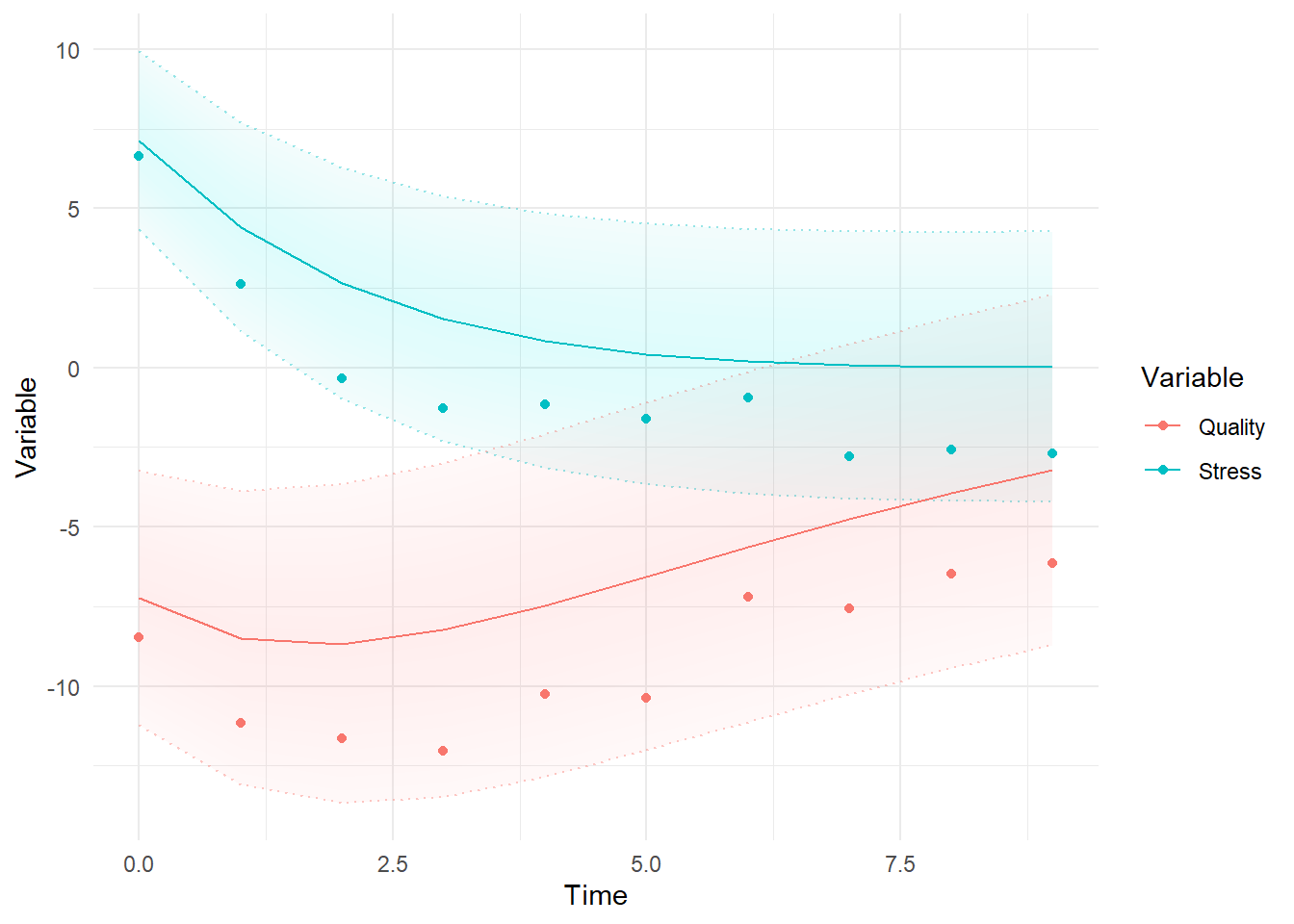
ctStanDiscretePars(fit,plot=T)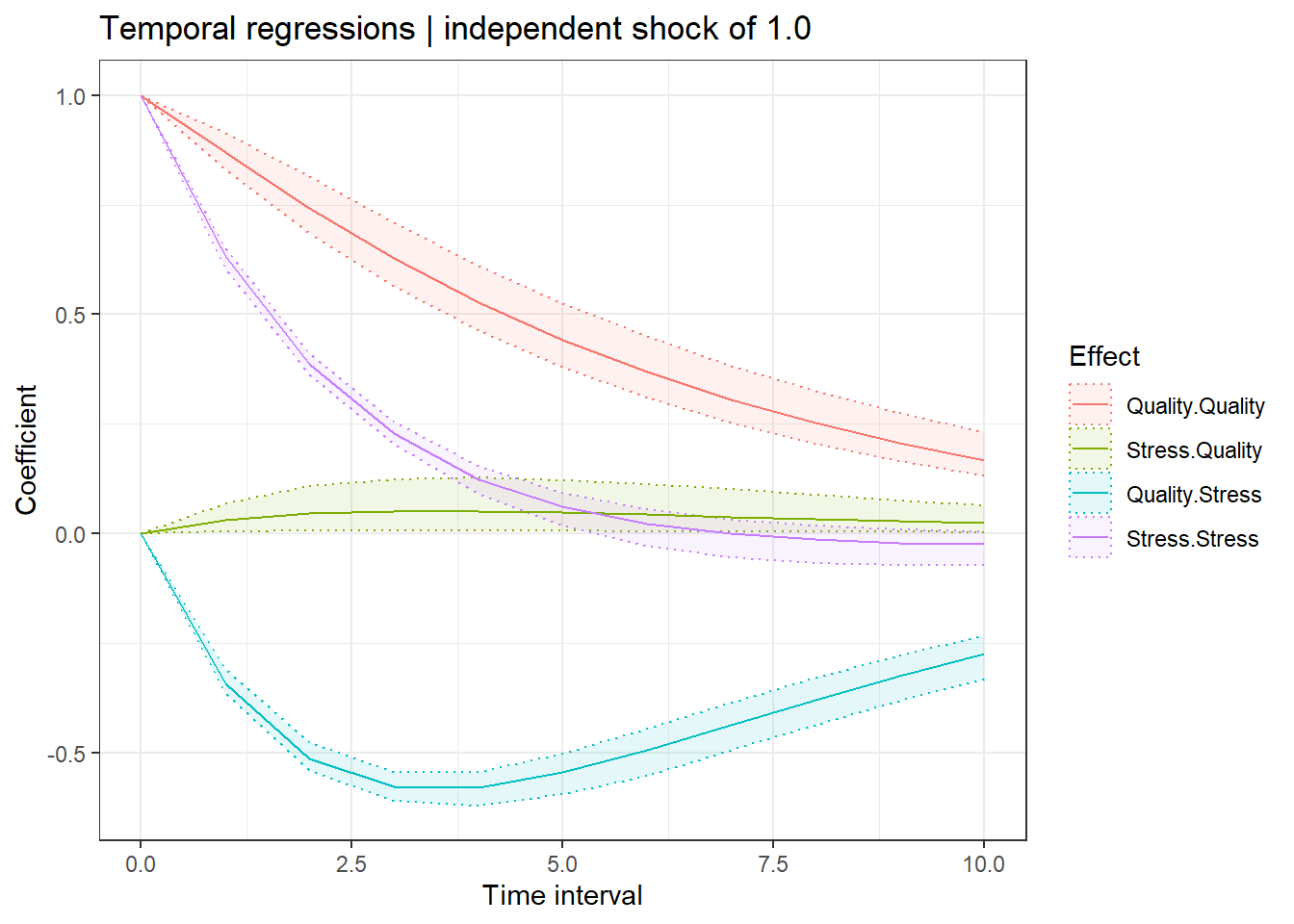
The previous plot shows the discrete-time regression coefficients for different lags – note that this is still a first order model (sometimes called lag 1), but because of the inclusion of measurement error and the possibility for missing data, higher lag effects are relevant. Put differently, the impact of a change at one point propagates further than the next observation, these plots show that propagation.
General summary information is found with the summary function, not shown here in full.
s <- summary(fit)
print(s$popmeans,digits=1) #basic system parameters
## mean sd 2.5% 50% 97.5%
## t0_stress 6.70 0.32 6.052 6.70 7.33
## t0_quality -7.80 0.60 -8.933 -7.77 -6.67
## a11 0.63 0.01 0.612 0.63 0.65
## a12 0.03 0.02 0.004 0.03 0.06
## a21 -0.34 0.01 -0.367 -0.34 -0.31
## a22 0.87 0.02 0.829 0.87 0.91
## diff11 0.63 0.03 0.579 0.63 0.68
## diff21 1.00 0.00 1.000 1.00 1.00
## diff22 0.82 0.04 0.748 0.82 0.89
## merr_stress 0.48 0.02 0.439 0.48 0.53
## merr_quality 0.53 0.03 0.470 0.53 0.60
## mi_stress 0.44 0.35 -0.237 0.43 1.13
## mi_quality 0.57 0.63 -0.613 0.54 1.82Amongst other things, from the above we can read off that the confidence interval for the a21 parameter – the 2nd row and 1st column of the temporal effects (DRIFT) matrix, representing the cross regression coefficient for the effect of the 1st process (stress) on the 2nd process (quality), is negative and zero is well outside the 95% confidence interval.
print(s$rawpopcorr,digits=2) #correlations between random effect parameters
## mean sd 2.5% 50% 97.5% z
## t0_quality__t0_stress 0.82 0.088 0.58 0.84 0.93 9.2
## mi_stress__t0_stress -0.73 0.064 -0.83 -0.74 -0.58 -11.4
## mi_quality__t0_stress -0.62 0.090 -0.77 -0.63 -0.42 -6.9
## mi_stress__t0_quality -0.50 0.106 -0.68 -0.51 -0.27 -4.7
## mi_quality__t0_quality -0.61 0.082 -0.76 -0.62 -0.44 -7.5
## mi_quality__mi_stress 0.91 0.024 0.85 0.91 0.95 38.4Here, we can see that the initial levels (t0), and long term levels (mi, for the manifest intercept) all correlate, while correlations between initial levels and long term levels appear negative.
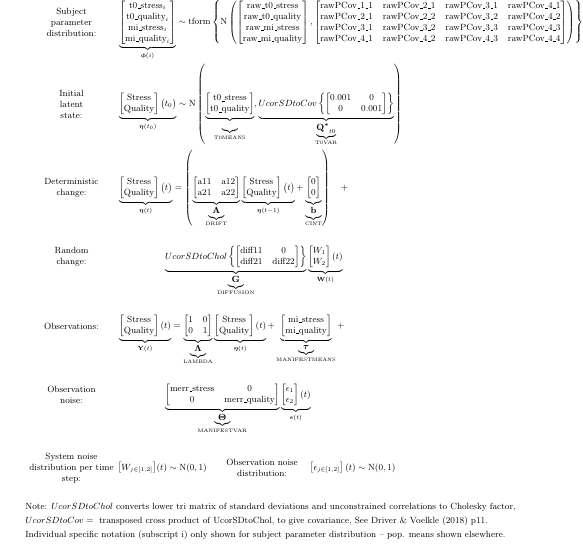
The fitted estimates can be seen in matrix representation too:
Predictive Checks
We can generate random samples of data based on our parameter estimates and compare these to our original data. The following is but one example of many possible checks, showing how the original data (in black) compares to the distribution of generated data (in red) over time, with means shown by the thicker dashed and solid lines. Based on only this plot, our estimated model looks like a very good match for the data. Which is not surprising because this is a toy example.
library(data.table)
fit <- ctStanGenerateFromFit(fit, nsamples=200, #add generated data to the fit object
fullposterior = F, cores = 2)
gendat <- as.data.table(fit$generated$Y) #extract generated data
gendat[,row:=as.integer(row)] #set the row column type appropriately
truedat <- melt(data.table(row=1:nrow(dat),dat), #get original data in melted form, ready for merging
measure.vars = c('Stress','Stress2','Quality'),variable.name = 'V1',value.name = 'TrueValue')
gendat <- merge(gendat,truedat,by = c('row','V1')) #merge original and melted data, ready for plotting
library(ggplot2)
ggplot(gendat,aes(y=value,x=time))+
stat_summary(fun.data=mean_sdl,geom='ribbon',alpha=.3,fill='red')+
stat_summary(aes(y = value), fun.y=mean, colour="red", geom="line",size=1)+
stat_summary(aes(y = TrueValue), fun.y=mean, geom="line",linetype='dashed',size=1)+
geom_line(aes(y=TrueValue,group=id),alpha=.2)+
facet_wrap(vars(V1),scales = 'free')+
theme_bw()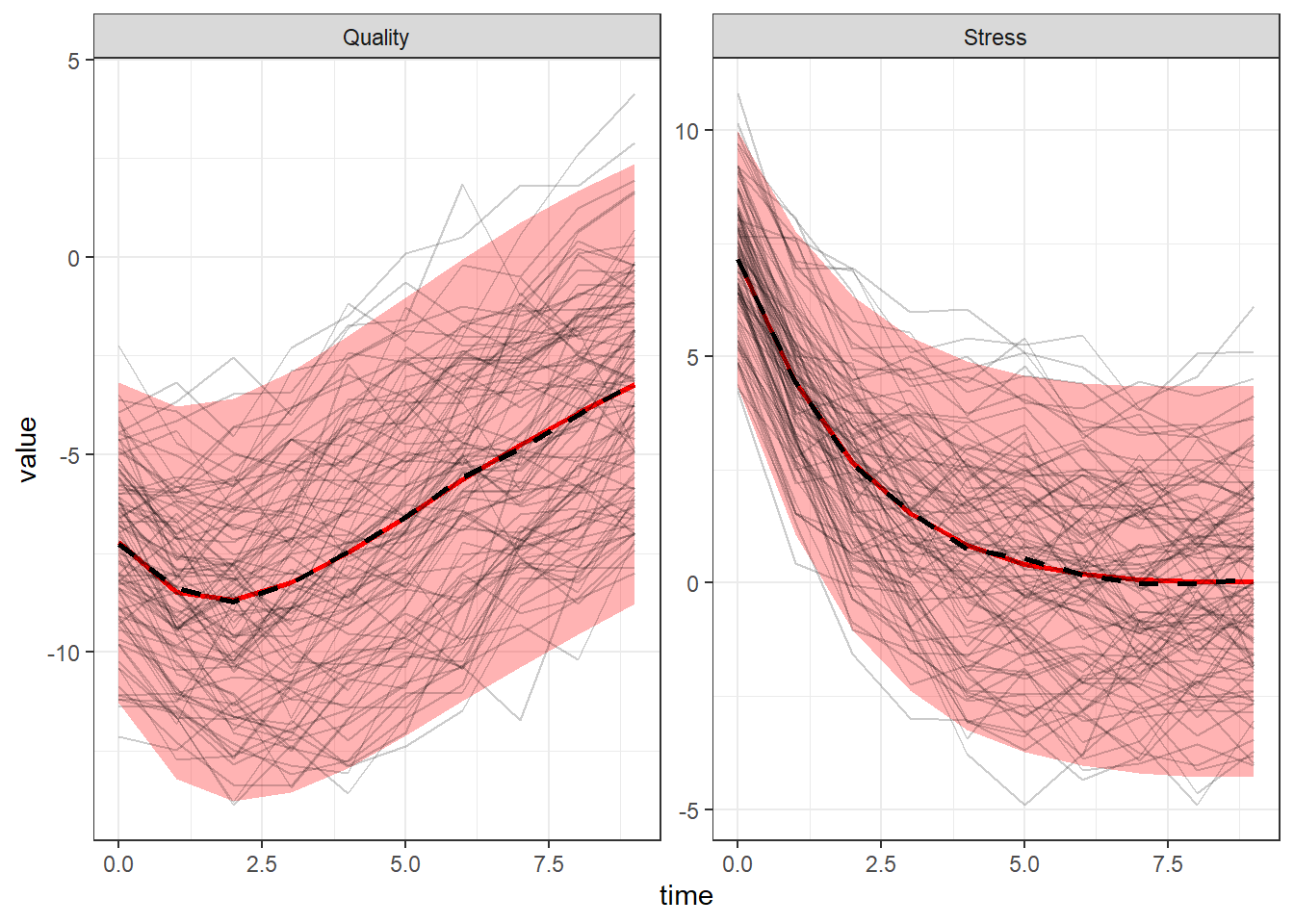
Residual Checks
We can extract the residuals and or standardised residuals from the model fit using the ctKalman function, then check for autocorrelation in the residuals – correlated residuals imply there is information left that our model did not leverage, meaning that our model could be improved. Here we show the autocorrelation using a method that treats time continuously, as well as the standard discrete-time approach in R. Here again our model is doing fine, the residual autocorrelation is approximately zero, there is no information remaining from the earlier time points to predict the latter.
k=ctKalman(fit,subjects=unique(dat$id)) #get residuals and various estimates
k=k[k$Element %in% 'errstdprior',] #remove everything except the residuals
k=dcast(data = data.table(k),formula = formula('Subject + Time ~ Row')) #cast to wider format
#continuous time auto and cross correlation
library(expct) #devtools::install_github("ryanoisin/expct")
expct(dataset = k,Time = 'Time',ID = 'Subject',outcome=c('Quality'),
plot_show = T,Tpred = 1:10,k = 5) #autocorrelation of residuals 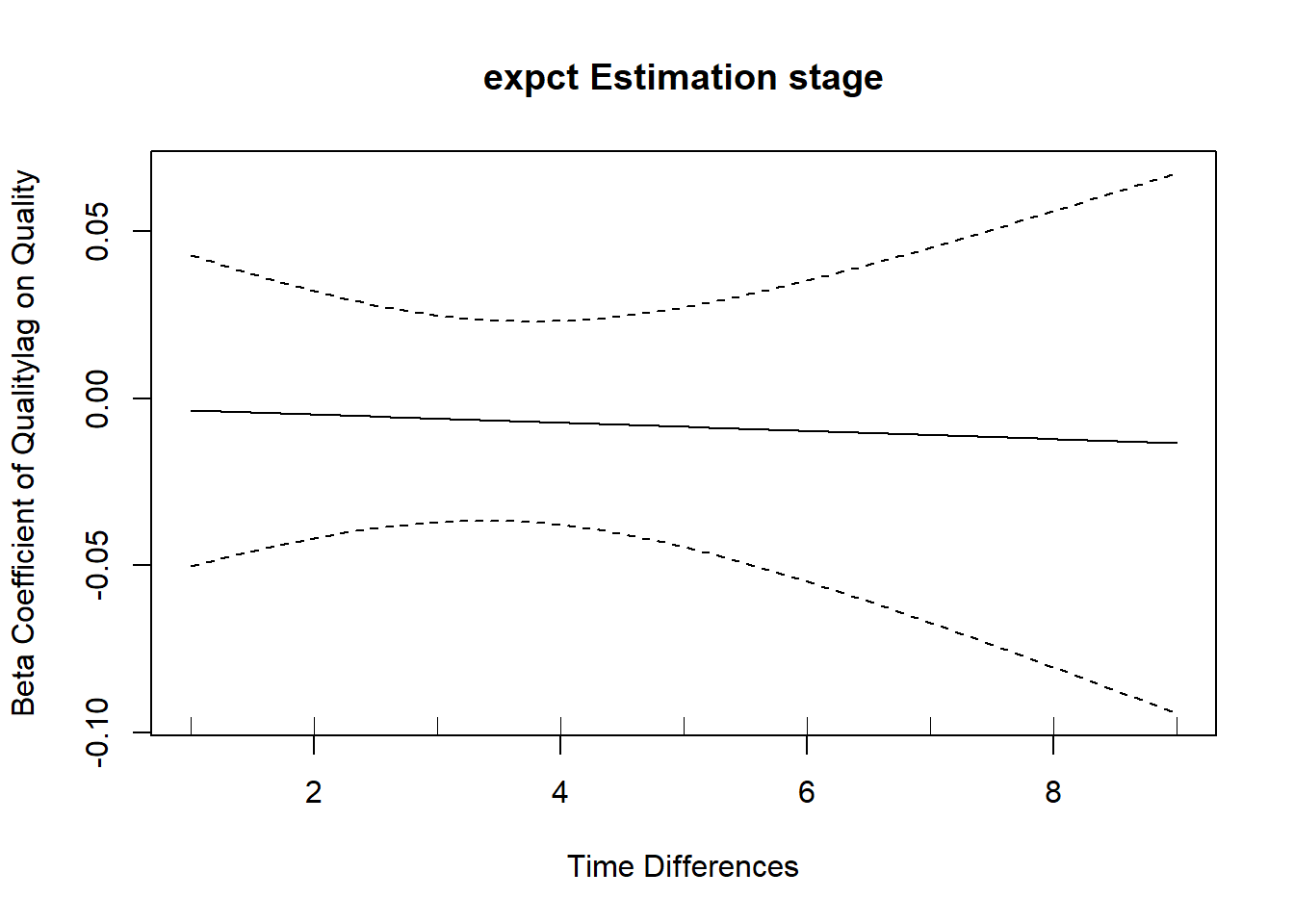
#discrete time auto correlation
acf(k$Quality,lag.max=10,plot=TRUE)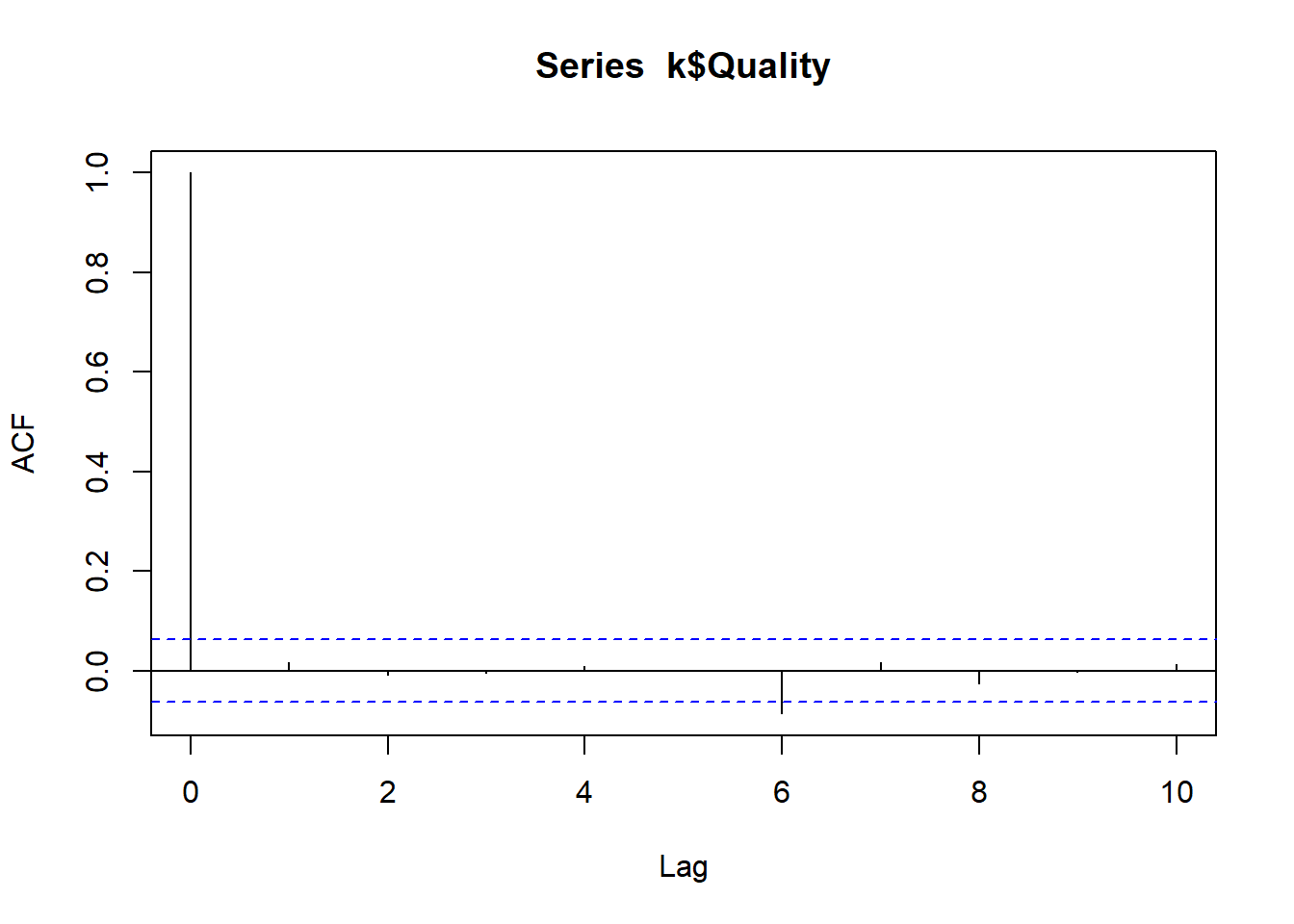
Factor / Measurement Models
Extending to a factor model formulation, using the additional measurement of stress we obtained, looks as follows, this time relying heavily on ctsem defaults for the free parameter specifications:
factormodel <- ctModel(type = 'standt',
manifestNames = c('Stress','Stress2','Quality'),
LAMBDA = matrix(c( #factor loadings / measurement structure
1,0,
'stressLoading',0, #now with estimated factor loading for Stress2 variable
0,1),ncol=2))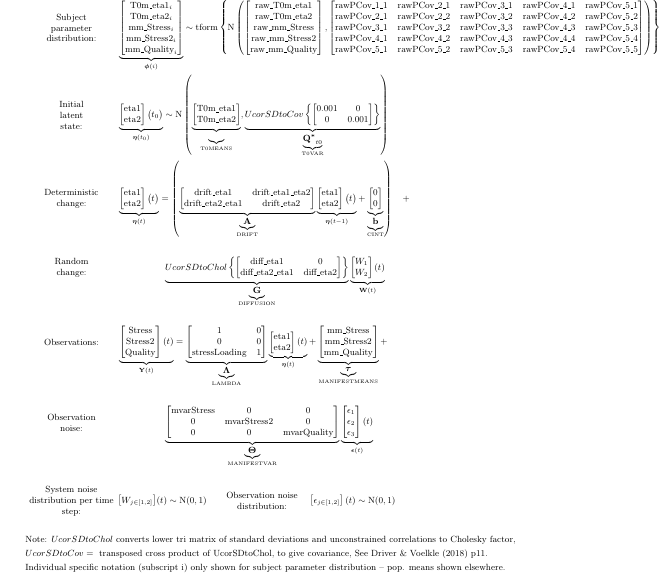
If we wanted to allow for stable individual variation only in the latent processes, and not in the measurement intercept, we would need to free the latent, or continuous, intercept (CINT, because it is added at each step), restrict all but one of the manifest intercepts (MANIFESTMEANS) to zero, and remove the individual variation from the estimated manifest intercept. Like this:
factormodellatent <- ctModel(type = 'standt',
manifestNames = c('Stress','Stress2','Quality'),
LAMBDA = matrix(c( #factor loadings / measurement structure
1,0,
'stressLoading',0, #now with estimated factor loading for Stress2 variable
0,1), ncol=2),
CINT=c('cintStress','cintQuality'),
MANIFESTMEANS=c(0,'mintStress2||FALSE',0)) #note the two || and FALSE, disabling individual variation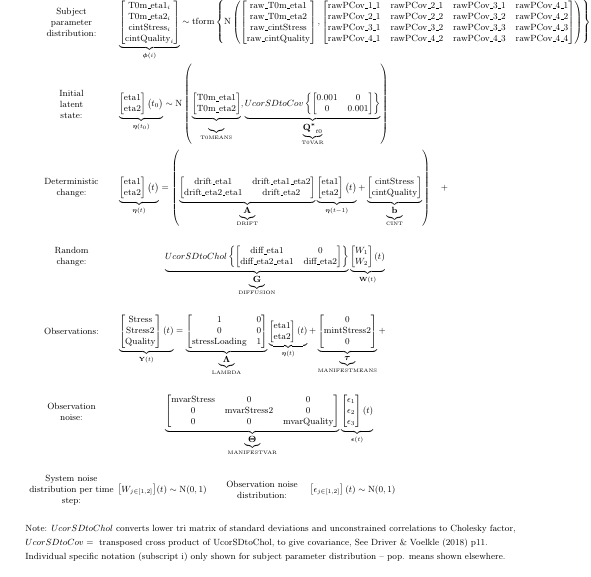
Want to account for heterogeneity in the model parameters without going wild and adding random effects (via the ||TRUE notation) to everything? Including the initial values and or mean of the observations as moderators of the parameters is one possibility. Here we first compute initial and mean values for our dataset, then include them as time independent predictors (covariate moderators). We also allow for random effects in the free factor loading – maybe not totally sensible in this case but simply as a demonstration how to add random effects to other parameters.
require(data.table)
dat <- data.table(dat)
dat[,initialStress:=mean(c(Stress[1],Stress2[1])),by=id]
dat[,meanStress:=mean(c(Stress,Stress2)),by=id]
head(dat)
factormodellatentmod<- ctModel(type = 'standt',
TIpredNames = c('initialStress','meanStress'),
tipredDefault = TRUE, #by default, moderate all parameters
CINT=c('cintStress||||meanStress', #only moderate cintStress using meanStress
'cintQuality||||initialStress, meanStress'), #explicitly moderate via both covariates
manifestNames = c('Stress','Stress2','Quality'),
LAMBDA = matrix(c( #factor loadings / measurement structure
1,0,
'stressLoading||TRUE',0, #estimated factor loading for Stress2 variable, now as random effect
0,1), ncol=2),
MANIFESTMEANS=c(0,'mintStress2||FALSE',0)) #note the two || and FALSE, disabling individual variationThe matrices for individual differences get pretty big now, zoom in if you like…